Intramolecular interaction analysis of twenty-seven benzothiazole derivatives with CDK9 using a theoretical model
DOI:
https://doi.org/10.14295/bjs.v4i12.798Keywords:
cancer, benzothiazole, CDK9, 3ocb proteinAbstract
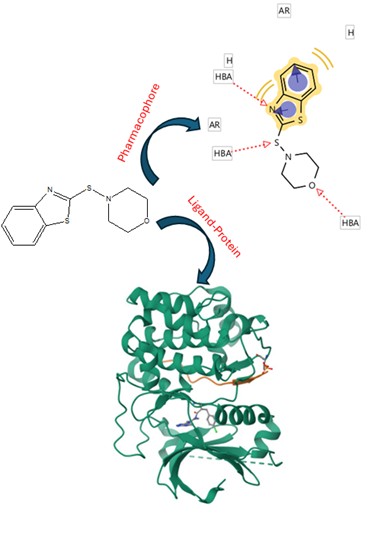
There are studies indicating that some drugs can regulate cancer cell growth through CDK9 inhibition. This study aimed to evaluate the possibility of twenty-seven benzothiazole analogs interacting with CDK9 using the 3ocb protein as a theoretical tool. In addition, the fedracib, KB-0742, and N-vinylpyrrolidone drugs were used as controls in the DockingServer program. The results showed different amino acid residues involved in the docking of benzothiazole derivatives (1-27) with the 3ocb protein surface compared to the controls. Other data displayed that the inhibition constant (Ki) was lower for compounds 1, 4, 7, 9, 11, 13-15, 17, 19-21, 22, 24, and 26 compared to KB-0742 and N-Vinylpyrrolidone. All this data indicate that these benzothiazole derivatives might have a higher affinity for the 3ocb protein surface, and this phenomenon could be translated as a CDK9 inhibition, resulting in a decrease in cancer cell growth.
References
Anshabo, A., Milne, R., Wang, S., & Albrecht, H. (2021). CDK9: a comprehensive review of its biology and its role as a potential target for anti-cancer agents. Frontiers in Oncology, 11, 678559. https://doi.org/10.3389/fonc.2021.678559 DOI: https://doi.org/10.3389/fonc.2021.678559
Askerova, U. (2023). Prediction of acute toxicity for (Z)-3-(2-phenylhydrazinylidene) benzofuran-2 (3H)-one and its derivatives for rats using GUSAR program. New Materials, Compounds and Applications, 7(1), 50-56.
Azzam, R., & Elgemeie, G. (2023). Purine analogs: synthesis, evaluation and molecular dynamics of pyrazolopyrimidines based benzothiazole as anticancer and antimicrobial CDK inhibitors. Nucleosides, Nucleotides & Nucleic Acids, 42(1), 77-104. https://doi.org/10.1080/15257770.2022.2109169 DOI: https://doi.org/10.1080/15257770.2022.2109169
Bakchi, B., Krishna, A., Sreecharan, E., Ganesh, V., Niharika, M., Maharshi, S., & Shaik, A. B. (2022). An overview on applications of SwissADME web tool in the design and developmentof anticancer, antitubercular and antimicrobial agents: a medicinal chemist's perspective. Journal of Molecular Structure, 1259, 132712. https://doi.org/10.1016/j.molstruc.2022.132712 DOI: https://doi.org/10.1016/j.molstruc.2022.132712
Banerjee, P., Eckert, A., Schrey, A., & Preissner, R. (2018). ProTox-II: a webserver for the prediction of toxicity of chemicals. Nucleic Acids Research, 46(W1), W257-W263. https://doi.org/10.1093/nar/gky318 DOI: https://doi.org/10.1093/nar/gky318
Barili, V., Ambrosini, E., Bortesi, B., Minari, R., De Sensi, E., Cannizzaro, I. R., & Pellegrino, B. (2024). Genetic basis of breast and ovarian cancer: approaches and lessons learnt from three decades of inherited predisposition testing. Genes, 15(2), 219. https://www.mdpi.com/2073-4425/15/2/219# DOI: https://doi.org/10.3390/genes15020219
Baroni M., Cruciani, G., Sciabola, S., Perruccio, F., Mason, J. (2007). A common reference framework for analyzing/comparing proteins and ligands. Fingerprints for ligands and proteins (FLAP): Theory and application. Journal of Chemical Information and Modeling, 47(2), 279-294. https://doi.org/ 10.1021/ci600253e DOI: https://doi.org/10.1021/ci600253e
Boffo, S., Damato, A., Alfano, L., & Giordano, A. (2018). CDK9 inhibitors in acute myeloid leukemia. Journal of Experimental & Clinical Cancer Research, 37(1), 36. DOI: https://doi.org/10.1186/s13046-018-0704-8
Çakmak, C., & Uğurluoğlu, Ö. (2024). The effects of patient-centered communication on patient engagement, health-related quality of life, service quality perception and patient satisfaction in patients with cancer: a cross-sectional study in Türkiye. Cancer Control, 31, 10732748241236327. https://doi.org/10.1177/10732748241236327 DOI: https://doi.org/10.1177/10732748241236327
Chen, I., & Foloppe, N. (2008). Conformational sampling of druglike molecules with MOE and catalyst: implications for pharmacophore modeling and virtual screening. Journal of Chemical Information and Modeling, 48(9), 1773-1791. DOI: https://doi.org/10.1021/ci800130k
Chen, R., Wierda, W. G., Chubb, S., Hawtin, R., Fox, J., Keating, M., & Plunkett, W. (2009). Mechanism of action of SNS-032, a novel cyclin-dependent kinase inhibitor, in chronic lymphocytic leukemia. Blood, The Journal of the American Society of Hematology, 113(19), 4637-4645. DOI: https://doi.org/10.1182/blood-2008-12-190256
Chen, Z., Wang, Z., Pang, J., Yu, Y., Bieerkehazhi, S., Lu, J., & Yang, J. (2016). Multiple CDK inhibitor dinaciclib suppresses neuroblastoma growth via inhibiting CDK2 and CDK9 activity. Scientific Reports, 6(1), 29090. DOI: https://doi.org/10.1038/srep29090
Deep, A., Marwaha, R., Marwaha, M., Nandal, R., & Sharma, A. K. (2018). Flavopiridol as cyclin dependent kinase (CDK) inhibitor: a review. New Journal of Chemistry, 42(23), 18500-18507.https://doi.org/10.1039/C8NJ04306J DOI: https://doi.org/10.1039/C8NJ04306J
Di Muzio, E. Toti, D., & Polticelli, F. (2017). DockingApp: a user friendly interface for facilitated docking simulations with AutoDock Vina. Journal of Computer-Aided Molecular Design, 31, 213-218 DOI: https://doi.org/10.1007/s10822-016-0006-1
Dixit, J., Gupta, N., Kataki, A., Roy, P., Mehra, N., Kumar, L., & Prinja, S. (2024). Health-related quality of life and its determinants among cancer patients: evidence from 12,148 patients of Indian database. Health and Quality of Life Outcomes, 22(1), 26. DOI: https://doi.org/10.1186/s12955-024-02227-0
Dixon, S., Smondyrev, A., Knoll, E., Rao, S., Shaw, D., & Friesner, R. (2006). PHASE: A new engine for pharmacophore perception, 3D QSAR model development, and 3D database screening: 1. Methodology and preliminary results. Journal of Computer-Aided Molecular Design, 20(10-11), 647-671. https://doi.org/10.1007/s10822-006-9087-6 DOI: https://doi.org/10.1007/s10822-006-9087-6
Figueroa-Valverde, L., Diaz-Cedillo, F., Rosas-Nexticapa, M., Cervantes-Ortega, C., Alvarez-Ramirez, M., Mateu-Armand, V., & Lopez-Ramos, M. (2023). Analysis of Interaction between Twenty-Seven Pyrimidinone Derivatives with XIAP Using a Theoretical Model. Clinical Cancer Investigation Journal, 12(3), 13-18. https://doi.org/10.51847/2bWWpF0Bdl Khedr, M. A., Zaghary, W. A., Elsherif, G. E., DOI: https://doi.org/10.51847/2bWWpF0Bdl
Figueroa-Valverde, L., Rosas-Nexticapa, M., Alvarez-Ramirez, M., Aguilar-Sanchez, E., Mateu-Armad, M. V., & Bonilla-Zavaleta, E. (2024). Interaction of some chalcone derivatives with calcium channels using a theoretical model. Brazilian Journal of Science, 3(11), 1-15. https://doi.org/10.14295/bjs.v3i11.658 DOI: https://doi.org/10.14295/bjs.v3i11.658
Franco, L., Morales, F., Boffo, S., & Giordano, A. (2018). CDK9: A key player in cancer and other diseases. Journal of Cellular Biochemistry, 119(2), 1273-1284. https://doi.org/10.1002/jcb.26293 DOI: https://doi.org/10.1002/jcb.26293
Habib, I., Chohan, T., Chohan, T., Batool, F., Khurshid, U., Khursheed, A., & Saleem, H. (2024). Integrated computational approaches for designing potent pyrimidine-based CDK9 inhibitors: 3D-QSAR, docking, and molecular dynamics simulations. Computational Biology and Chemistry, 108, 108003. https://doi.org/10.1016/j.compbiolchem.2023.108003 DOI: https://doi.org/10.1016/j.compbiolchem.2023.108003
Halgren. (1998). Merck molecular force field. I. Basis, form, scope, parametrization, and performance of MMFF94. Journal of Computational Chemistry, 17(5-6), 490-519. DOI: https://doi.org/10.1002/(SICI)1096-987X(199604)17:5/6<490::AID-JCC1>3.0.CO;2-P
Housini, M., Dariya, B., Ahmed, N., Stevens, A., Fiadjoe, H., Nagaraju, G. P., & Basha, R. (2024). Colorectal cancer: Genetic alterations, novel biomarkers, current therapeutic strategies and clinical trials. Gene, 892, 147857. https://doi.org/10.1016/j.gene.2023.147857 DOI: https://doi.org/10.1016/j.gene.2023.147857
Hussain, A., Verma, C., & Chouhan, U. (2017). Identification of novel inhibitors against Cyclin Dependent Kinase 9/Cyclin T1 complex as: Anti cancer agent. Saudi Journal of Biological Sciences, 24(6), 1229-1242.https://doi.org/10.1016/j.sjbs.2015.10.003 DOI: https://doi.org/10.1016/j.sjbs.2015.10.003
Ionescu, A., Anghel, A., Antone-Iordache, I., Atasiei, D., Anghel, C., Barnonschi, A., & Lișcu, H. (2024). Assessing the impact of organ failure and metastases on quality of life in breast cancer patients: a prospective study based on utilizing EORTC QLQ-C30 and EORTC QLQ-BR45 questionnaires in Romania. Journal of Personalized Medicine, 14(2), 214. https://www.mdpi.com/2075-4426/14/2/214# DOI: https://doi.org/10.3390/jpm14020214
Irfan, A., Batool, F., Zahra Naqvi, S., Islam, A., Osman, S., Nocentini, A., & Supuran, C. T. (2020). Benzothiazole derivatives as anticancer agents. Journal of Enzyme Inhibition and Medicinal Chemistry, 35(1), 265-279.https://doi.org/10.1080/14756366.2019.1698036 DOI: https://doi.org/10.1080/14756366.2019.1698036
Kini, S., Swain, S., & Gandhi, A. (2007). Synthesis and evaluation of novel benzothiazole derivatives against human cervical cancer cell lines. Indian Journal of Pharmaceutical Sciences, 69(1), 46-50. DOI: https://doi.org/10.4103/0250-474X.32107
Koes, D., Camacho, C. (2011). Pharmer: Efficient and exact pharmacophore search. Journal of Chemical Information and Modeling, 51(6), 1307-1314. https://doi.org/ 10.1021/ci200097m DOI: https://doi.org/10.1021/ci200097m
Kok, S., Gambari, R., Chui, C., Yuen, M., Lin, E., Wong, R., & Chan, A. (2008). Synthesis and anti-cancer activity of benzothiazole containing phthalimide on human carcinoma cell lines. Bioorganic & Medicinal Chemistry, 16(7), 3626-3631. https://doi.org/10.1016/j.bmc.2008.02.005 DOI: https://doi.org/10.1016/j.bmc.2008.02.005
Lee, D., & Zeidner, J. (2019). Cyclin-dependent kinase (CDK) 9 and 4/6 inhibitors in acute myeloid leukemia (AML): a promising therapeutic approach. Expert Opinion on Investigational Drugs, 28(11), 989-1001. https://doi.org/10.1080/13543784.2019.1678583 DOI: https://doi.org/10.1080/13543784.2019.1678583
Lücking, U., Scholz, A., Lienau, P., Siemeister, G., Kosemund, D., Bohlmann, R., & Brands, M. (2017). Identification of atuveciclib (BAY 1143572), the first highly selective, clinical PTEFb/CDK9 inhibitor for the treatment of cancer. ChemMedChem, 12(21), 1776-1793.https://doi.org/10.1002/cmdc.201700447 DOI: https://doi.org/10.1002/cmdc.201700447
Liu, H., Guo, Z., & Wang, P. (2024). Genetic expression in cancer research: challenges and complexity. Gene reports, 37,102042. https://doi.org/10.1016/j.genrep.2024.102042 DOI: https://doi.org/10.1016/j.genrep.2024.102042
Ma, H., Seebacher, N., Hornicek, F., & Duan, Z. (2019). Cyclin-dependent kinase 9 (CDK9) is a novel prognostic marker and therapeutic target in osteosarcoma. EBioMedicine, 39, 182-193. DOI: https://doi.org/10.1016/j.ebiom.2018.12.022
Mandal, R., Becker, S., & Strebhardt, K. (2021). Targeting CDK9 for anti-cancer therapeutics. Cancers, 13(9), 2181. https://www.mdpi.com/2072-6694/13/9/2181# DOI: https://doi.org/10.3390/cancers13092181
Mohamed, L., Taher, A., Rady, G., Ali, M., & Mahmoud, A. (2017). Synthesis and cytotoxic activity of certain benzothiazole derivatives against human MCF‐7 cancer cell line. Chemical Biology & Drug Design, 89(4), 566-576.https://doi.org/10.1111/cbdd.12879 DOI: https://doi.org/10.1111/cbdd.12879
Morales, F., & Giordano, A. (2016). Overview of CDK9 as a target in cancer research. Cell Cycle, 15(4), 519-527. https://doi.org/10.1080/15384101.2016.1138186 DOI: https://doi.org/10.1080/15384101.2016.1138186
Morris, M., Goodsell, D., Hallyday, R., Huey, R., Hart, W., Belew, R., & Olson, A. (1998). Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. Journal of Computational Chemistry, 19(14), 1639-1662. https://doi.org/10.1002/(SICI)1096-987X(19981115)19:14%3C1639:AID-JCC10%3E3.0. CO;2-B DOI: https://doi.org/10.1002/(SICI)1096-987X(19981115)19:14<1639::AID-JCC10>3.0.CO;2-B
Noblejas-López, M., Gandullo-Sánchez, L., Galán-Moya, E., López-Rosa, R., Tébar-García, D., Nieto-Jiménez, C., & Ocaña, A. (2022). Antitumoral activity of a CDK9 PROTAC compound in HER2-positive breast cancer. International Journal of Molecular Sciences, 23(10), 5476. https://doi.org/10.3390/ijms23105476 DOI: https://doi.org/10.3390/ijms23105476
Parvathareddy, S., Siraj, A., Masoodi, T., Annaiyappanaidu, P., Al-Badawi, I., Al-Dayel, F., & Al-Kuraya, K. (2021). Cyclin-dependent kinase 9 (CDK9) predicts recurrence in Middle Eastern epithelial ovarian cancer. Journal of Ovarian Research, 14(1), 69. DOI: https://doi.org/10.1186/s13048-021-00827-8
Pathak, N., Rathi, E., Kumar, N., Kini, S., & Rao, C. (2020). A review on anticancer potentials of benzothiazole derivatives. Mini Reviews in Medicinal Chemistry, 20(1), 12-23. https://doi.org/10.2174/1389557519666190617153213 DOI: https://doi.org/10.2174/1389557519666190617153213
Plewczynski, D., Philips, A., Grotthuss, M., RychlewskiL., & Ginalski, K. (2014). HarmonyDOCK: the structural analysis of poses in protein-ligand docking. Journal of Computational Biology, 21(3), 247-256. https://doi.org/10.1089/cmb.2009.0111 DOI: https://doi.org/10.1089/cmb.2009.0111
Polier, G., Ding, J., Konkimalla, B., Eick, D., Ribeiro, N., Köhler, R., & Li-Weber, M. (2011). Wogonin and related natural flavones are inhibitors of CDK9 that induce apoptosis in cancer cells by transcriptional suppression of Mcl-1. Cell Death & Disease, 2(7), e182-e182. DOI: https://doi.org/10.1038/cddis.2011.66
Riniker, S., Christ, C., Hansen, H., Hünenberger, P., Oostenbrink, C., Steiner, D., & Van-Gunsteren, W. (2011). Calculation of relative free energy for ligand-protein binding, solvation, and conformational transitions using the GROMOS software. The Journal of Physical Chemistry B, 115(46), 13570-13577. https://doi.org/10.1021/jp204303a DOI: https://doi.org/10.1021/jp204303a
Saikat, A., Al-Khafaji, K., Akter, H., Choi, J., Hasan, M., & Lee, S. (2022). Nature-Derived Compounds as Potential Bioactive Leads against CDK9-Induced Cancer: Computational and Network Pharmacology Approaches. Processes, 10(12), 2512. https://doi.org/ 10.3390/pr10122512 DOI: https://doi.org/10.3390/pr10122512
Sarhadi, V. K., & Armengol, G. (2022). Molecular biomarkers in cancer. Biomolecules, 12, 1021. https://doi.org/10.3390/biom12081021 DOI: https://doi.org/10.3390/biom12081021
Shweta, M., & Rashmi, D. (2019). In-vitro ADME studies of TUG-891, a GPR-120 inhibitor using Swiss ADME predictor. Journal of Drug Delivery and Therapeutics, 9(2-S), 266-369. DOI: https://doi.org/10.22270/jddt.v9i2-s.2710
Singh, P., Kumar, V., Jung, T., Lee, J., Lee, K., & Hong, J. (2024). Uncovering potential CDK9 inhibitors from natural compound databases through docking-based virtual screening and MD simulations. Journal of Molecular Modeling, 30(8), 267. DOI: https://doi.org/10.1007/s00894-024-06067-z
Solis, F., & Wets, R. (1981). Minimization by Random Search Techniques. Mathematics of Operations Research, 6(1), 19-30. https://doi.org/10.1287/moor.6.1.19 DOI: https://doi.org/10.1287/moor.6.1.19
Stankovic, S., Shekari, S., Huang, Q. Q., Gardner, E. J., Ivarsdottir, E. V., Owens, N. D., & Murray, A. (2024). Genetic links between ovarian ageing, cancer risk and de novo mutation rates. Nature, 633(8030), 608-614. DOI: https://doi.org/10.1038/s41586-024-07931-x
Sushko, I., Salmina, E., Potemkin, V., Poda, G., & Tetko, I. (2012). ToxAlerts: a web server of structural alerts for toxic chemicals and compounds with potential adverse reactions. Journal of Chemical Information and Modeling, 52(8), 2310-2316. https://doi.org/10.1021/ci300245q DOI: https://doi.org/10.1021/ci300245q
Trosset, J., & Scheraga, H. (1999). PRODOCK: software package for protein modeling and docking. Journal of Computational Chemistry, 20(4), 412-427. https://doi.org/10.1002/(SICI)1096-987X(199903)20:4%3C412:AID-JCC3%3E3.0.CO;2-N DOI: https://doi.org/10.1002/(SICI)1096-987X(199903)20:4<412::AID-JCC3>3.0.CO;2-N
Uremis, N., Uremis, M., Tolun, F., Ceylan, M., Doganer, A., & Kurt, A. (2017). Synthesis of 2-substituted benzothiazole derivatives and their in vitro anticancer effects and antioxidant activities against pancreatic cancer cells. Anticancer Research, 37(11), 6381-6389. DOI: https://doi.org/10.21873/anticanres.12091
Xie, S., Jiang, H., Zhai, X., Wei, F., Wang, S., Ding, J., & Chen, Y. (2016). Antitumor action of CDK inhibitor LS-007 as a single agent and in combination with ABT-199 against human acute leukemia cells. Acta Pharmacologica Sinica, 37(11), 1481-1489. DOI: https://doi.org/10.1038/aps.2016.49
Zhang, H., Huang, J., Chen, R., Cai, H., Chen, Y., He, S., & Wang, L. (2022). Ligand-and structure-based identification of novel CDK9 inhibitors for the potential treatment of leukemia. Bioorganic & Medicinal Chemistry, 72, 116994. https://doi.org/10.1016/j.bmc.2022.116994 DOI: https://doi.org/10.1016/j.bmc.2022.116994
Zhang, H., Pandey, S., Travers, M., Sun, H., Morton, G., Madzo, J., & Issa, J. (2018). Targeting CDK9 reactivates epigenetically silenced genes in cancer. Cell, 175(5), 1244-1258. DOI: https://doi.org/10.1016/j.cell.2018.09.051
Zhang, M., Xia, Y., Tan, Y., Xie, Z., & Li, J. (2024). Expression of CDK9 in Newly Diagnosed Patients with Acute Myeloid Leukemia and its Clinical Significance. Clinical Laboratory, 70(10). DOI: https://doi.org/10.7754/Clin.Lab.2024.240416
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2025 Marcela Rosas Nexticapa; Magdalena Alvarez-Ramirez, Maria Virginia Mateu-Armad, Regina Cauich-Carrillo

This work is licensed under a Creative Commons Attribution 4.0 International License.
Authors who publish with this journal agree to the following terms:
1) Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution License that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
2) Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the journal's published version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgement of its initial publication in this journal.
3) Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work.


