Acetylcholinesterase inhibitory potential of plant-based phenolics in the treatment of Alzheimer's disease: An in silico approach
DOI:
https://doi.org/10.14295/bjs.v4i10.769Keywords:
acetylcholinesterase, ADMET modeling, Alzheimer’s disease, molecular docking, molecular dynamicsAbstract
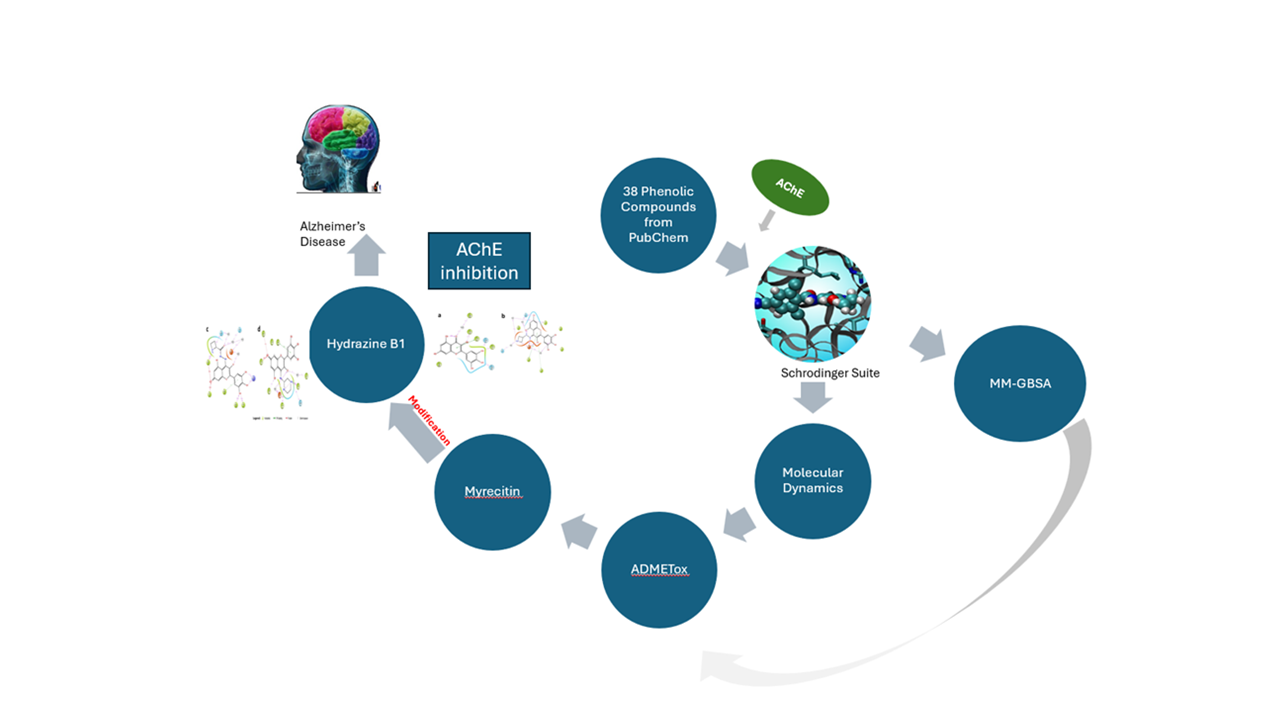
Alzheimer's disease is the most prevalent cause of dementia, accounting for more than seventy per cent of all the reported cases. Among the various treatment strategies, inhibiting the action of acetylcholinesterase that breaks down the neurotransmitter acetylcholine is the most common. In this report, thirty-eight phenolic compounds were retrieved from the PubChem database and screened in silico against acetylcholinesterase. Non-covalent molecular docking, molecular mechanics-generalized born surface area (MM-GBSA), and molecular dynamics (MD) were used to predict their binding mode, affinity, free energy, and the stability of the protein-ligand complex. These were followed by drug-likeness screening and a rigorous prediction of their absorption, distribution, metabolism, excretion, and toxicity (ADMET) parameters. Myricetin (-13.9 kcal/mol) was predicted to have the highest binding affinity among the phenolics, though lower than the bound donepezil (-16.3 kcal/mol). To increase the binding affinity of myricetin, it was modified via a Schiff base formation, which gave the hydrazine B-1 a binding affinity of -17.7 kcal/mol, higher than that of donepezil. The molecular dynamics simulation showed that the modified ligands have better stability than myricetin. The ADMET and drug-likeness studies showed that the top four phenolics and myricetin analogue derivatives could be further developed as potential drug candidates.
Keywords: Acetylcholinesterase, ADMET Modeling, Alzheimer’s disease, Molecular Docking, Molecular Dynamics
References
Alzheimer’s disease facts and figures. (2024). Alzheimer’s and Dementia, 20(5), 3708–3821. https://doi.org/10.1002/alz.13809 DOI: https://doi.org/10.1002/alz.13809
Abuzaid, H., Amin, E., Moawad, A., Usama Ramadan, Abdelmohsen, Hetta, M., & Mohammed1, R. (2020). Liquid Chromatography High-Resolution Mass Spectrometry Analysis, Phytochemical and Biological Study of Two Aizoaceae Plants Plants: A New Kaempferol Derivative from Trianthema portulacastrum L. Pharmacognosy Research, 10, 24–30. https://doi.org/10.4103/pr.pr
Adelusi, T. I., Adeyemi, R. O., Ashiru, M. A., Divine, U. C., Boyenle, I. D., Oyedele, A. Q. K., & Adewoye, I. M. (2023). Prediction of Antidiabetic Compounds in Curcuma longa – In vitro and In silico Investigations. Tropical Journal of Natural Product Research, 7(10), 4937–4944. https://doi.org/10.26538/tjnpr/v7i10.33 DOI: https://doi.org/10.26538/tjnpr/v7i10.33
Adelusi, T. I., Oyedele, A. Q. K., Monday, O. E., Boyenle, I. D., Idris, M. O., Ogunlana, A. T., Ayoola, A. M., Fatoki, J. O., Kolawole, O. E., David, K. B., & Olayemi, A. A. (2022). Dietary polyphenols mitigate SARS-CoV-2 main protease (Mpro)–Molecular dynamics, molecular mechanics, and density functional theory investigations. Journal of Molecular Structure, 1250, 131879. https://doi.org/10.1016/j.molstruc.2021.131879 DOI: https://doi.org/10.1016/j.molstruc.2021.131879
Adeoye, M. D., Oyebamiji, A. K., Ashiru, M. A., Adigun, R. A., Olalere, O. H., & Semire, B. (2022). Biological evaluation of selected metronidazole derivatives as anti-nitroreductase via in silico approach. Ecletica Quimica, 47(4), 27–36. https://doi.org/10.26850/1678-4618eqj.v47.4.2022.p27-36 DOI: https://doi.org/10.26850/1678-4618eqj.v47.4.2022.p27-36
Adigun, R. A., Malan, F. P., Balogun, M. O., & October, N. (2019). Tetrahydropyrimidinones/thiones stabilized by trifluoromethyl-containing β-diketones. Journal of Molecular Structure, 1202, 127281. https://doi.org/10.1016/j.molstruc.2019.127281 DOI: https://doi.org/10.1016/j.molstruc.2019.127281
Ashiru, M. A., Ogunyemi, S. O., Temionu, O. R., Ajibare, A. C., Cicero-Mfon, N. C., Ihekuna, O. A., Jagun, M. O., Abdulmumin, L., Adisa, Q. K., Asibor, Y. E., Okorie, C. J., Lawal, M. O., Babalola, M. O., Abdulrasaq, I. T., Salau, L. B.,
Olatunji, I. O., Bankole, M. A., Daud, A. B., & Adeyemi, A. O. (2023). Identification of EGFR inhibitors as potential agents for cancer therapy: pharmacophore-based modeling, molecular docking, and molecular dynamics investigations. Journal of Molecular Modeling, 29(5). https://doi.org/10.1007/s00894-023-05531-6 DOI: https://doi.org/10.1007/s00894-023-05531-6
Association, A. (2022). 2022 Alzheimer’s disease facts and figures. Alzheimer’s & Dementia, 18(4), 700-789. https://doi.org/10.1002/alz.12638 DOI: https://doi.org/10.1002/alz.12638
Babalola, M. O., Ashiru, M. A., Boyenle, I. D., Atanda, E. O., Oyedele, A.-Q. K., Dimeji, I. Y., Awodele, O., & Imaga, N. A. (2022). In vitro Analysis and Molecular Docking of Gas Chromatography-Mass Spectroscopy Fingerprints of Polyherbal Mixture Reveals Significant Antidiabetic Miture. Nigerian Journal of Experimental and Clinical Biosciences, 10(4), 105-115. https://doi.org/10.4103/njecp.njecp_15_22 DOI: https://doi.org/10.4103/njecp.njecp_15_22
Bai, Y. R., Seng, D. J., Xu, Y., Zhang, Y. D., Zhou, W. J., Jia, Y. Y., Song, J., He, Z. X., Liu, H. M., & Yuan, S. (2024). A comprehensive review of small molecule drugs approved by the FDA in 2023: Advances and prospects. In European Journal of Medicinal Chemistry, 276. https://doi.org/10.1016/j.ejmech.2024.116706 DOI: https://doi.org/10.1016/j.ejmech.2024.116706
Barak, D., Ordentlich, A., Stein, D., Yu, Q., Greig, N. H., & Shafferman, A. (2009). Accommodation of physostigmine and its analogues by acetylcholinesterase is dominated by hydrophobic interactions. Biochemical Journal, 417(1), 213-222. DOI: https://doi.org/10.1042/BJ20081276
Bhandari, R., Gyawali, S., Aryal, N., Gaire, D., Paudyal, K., Panta, A., Panth, P., Joshi, D. R., Rokaya, R. K., Aryal, P., & Pandey, J. (2021). Evaluation of phytochemical, antioxidant, and memory-enhancing activity of Garuga pinnata Roxb. Bark and Bryophyllum pinnatum (Lam) Oken. leaves. The Scientific World Journal, 2021, 1-7. https://doi.org/10.1155/2021/6649574 DOI: https://doi.org/10.1155/2021/6649574
Bhuia, M. S., Rahaman, M. M., Islam, T., Bappi, M. H., Sikder, M. I., Hossain, K. N., Akter, F., Al Shamsh Prottay, A., Rokonuzzman, M., Gürer, E. S., Calina, D., Islam, M. T., & Sharifi-Rad, J. (2023). Neurobiological effects of gallic acid: current perspectives. In Chinese Medicine (United Kingdom), 18(1). https://doi.org/10.1186/s13020-023-00735-7 DOI: https://doi.org/10.1186/s13020-023-00735-7
Dileep, K. V., Ihara, K., Mishima-Tsumagari, C., Kukimoto-Niino, M., Yonemochi, M., Hanada, K., Shirouzu, M., & Zhang, K. Y. J. (2022). Crystal structure of human acetylcholinesterase in complex with tacrine: Implications for drug discovery. International Journal of Biological Macromolecules, 210, 172-181. https://doi.org/10.1016/j.ijbiomac.2022.05.009 DOI: https://doi.org/10.1016/j.ijbiomac.2022.05.009
Efferth, T., Xu, A. L., & Lee, D. Y. W. (2019). Combining the wisdoms of traditional medicine with cutting-edge science and technology at the forefront of medical sciences. In Phytomedicine, 64. https://doi.org/10.1016/j.phymed.2019.153078 DOI: https://doi.org/10.1016/j.phymed.2019.153078
English, C. (2012). How the FDA forgot the evidence: the case of donepezil 23 mg. BMJ, 344. https://doi.org/10.1136/bmj.e1086 DOI: https://doi.org/10.1136/bmj.e1086
Ferreira, L. L. G., & Andricopulo, A. D. (2019). ADMET modeling approaches in drug discovery. Drug Discovery Today, 24(5), 1157-1165. DOI: https://doi.org/10.1016/j.drudis.2019.03.015
Genheden, S., & Ryde, U. (2015). The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opinion on Drug Discovery, 10(5), 449-461. DOI: https://doi.org/10.1517/17460441.2015.1032936
Gupta, G., Siddiqui, M. A., Khan, M. M., Ajmal, M., Ahsan, R., Rahaman, M. A., Ahmad, M. A., Arshad, M., & Khushtar, M. (2020). Current pharmacological trends on myricetin. Drug Research. DOI: https://doi.org/10.1055/a-1224-3625
Haider, M. K., Bertrand, H. O., & Hubbard, R. E. (2011). Predicting fragment binding poses using a combined MCSS MM-GBSA approach. Journal of Chemical Information and Modeling, 51(5), 1092-1105. https://doi.org/10.1021/ci100469n DOI: https://doi.org/10.1021/ci100469n
Hofer, T., & Perry, G. (2016). Nucleic acid oxidative damage in Alzheimer’s disease—explained by the hepcidin-ferroportin neuronal iron overload hypothesis? In Journal of Trace Elements in Medicine and Biology, 38, 1-9. https://doi.org/10.1016/j.jtemb.2016.06.005 DOI: https://doi.org/10.1016/j.jtemb.2016.06.005
Hosseini, F., Mohammadi-Khanaposhtani, M., Azizian, H., Ramazani, A., Tehrani, M. B., Nadri, H., Larijani, B., Biglar, M., Adibi, H., & Mahdavi, M. (2020). 4-Oxobenzo[d]1,2,3-triazin-pyridinium-phenylacetamide derivatives as new anti-Alzheimer agents: design, synthesis, in vitro evaluation, molecular modeling, and molecular dynamic study. Structural Chemistry, 31(3), 999-1012. https://doi.org/10.1007/s11224-019-01472-0 DOI: https://doi.org/10.1007/s11224-019-01472-0
Islam, B. ul, Jabir, N. R., & Tabrez, S. (2019). The role of mitochondrial defects and oxidative stress in Alzheimer’s disease. In Journal of Drug Targeting, 27(9), 932-942. https://doi.org/10.1080/1061186X.2019.1584808 DOI: https://doi.org/10.1080/1061186X.2019.1584808
Jabir, N. R., Rehman, M. T., Alsolami, K., Shakil, S., Zughaibi, T. A., Alserihi, R. F., Khan, M. S., AlAjmi, M. F., & Tabrez, S. (2021). Concatenation of molecular docking and molecular simulation of BACE-1, γ-secretase targeted ligands: in pursuit of Alzheimer’s treatment. Annals of Medicine, 53(1), 2332-2344. https://doi.org/10.1080/07853890.2021.2009124 DOI: https://doi.org/10.1080/07853890.2021.2009124
Jabir, N. R., Shakil, S., Tabrez, S., Khan, M. S., Rehman, M. T., & Ahmed, B. A. (2021). In silico screening of glycogen synthase kinase-3β targeted ligands against acetylcholinesterase and its probable relevance to Alzheimer’s disease. Journal of Biomolecular Structure and Dynamics, 39(14), 5083-5092. https://doi.org/10.1080/07391102.2020.1784796 DOI: https://doi.org/10.1080/07391102.2020.1784796
Kaus, J. W., Harder, E., Lin, T., Abel, R., McCammon, J. A., & Wang, L. (2015). How to deal with multiple binding poses in alchemical relative protein–ligand binding free energy calculations. Journal of Chemical Theory and Computation, 11(6), 2670-2679. DOI: https://doi.org/10.1021/acs.jctc.5b00214
Kehinde, O. A.-Q., Damilare, B. I., Ogunlana, A., Ayoola, A. M., Opeyemi Emmanuel, A., & Temitope Isaac, A. (2022). Inhibitors of α-glucosidase and angiotensin-converting enzyme in the treatment of type 2 diabetes and its complications: A review on in silico approach. Pharmaceutical and Biomedical Research, 8(4), 237-258. https://doi.org/10.32598/PBR.8.4.1052.1 DOI: https://doi.org/10.32598/PBR.8.4.1052.1
Khan, S., Hassan, M. I., Shahid, M., & Islam, A. (2023). Nature’s toolbox against tau aggregation: An updated review of current research. In Ageing Research Reviews, 87. https://doi.org/10.1016/j.arr.2023.101924 DOI: https://doi.org/10.1016/j.arr.2023.101924
Kiokias, S., Proestos, C., & Oreopoulou, V. (2020). Phenolic acids of plant origin-a review on their antioxidant activity in vitro (O/W emulsion systems) along with their in vivo health biochemical properties. In Foods, 9(4). https://doi.org/10.3390/foods9040534 DOI: https://doi.org/10.3390/foods9040534
Lu, C., Wu, C., Ghoreishi, D., Chen, W., Wang, L., Damm, W., Ross, G. A., Dahlgren, M. K., Russell, E., & Von Bargen, C. D. (2021). OPLS4: Improving force field accuracy on challenging regimes of chemical space. Journal of Chemical Theory and Computation. DOI: https://doi.org/10.1021/acs.jctc.1c00302
Madav, Y., Wairkar, S., & Prabhakar, B. (2019). Recent therapeutic strategies targeting beta amyloid and tauopathies in Alzheimer’s disease. In Brain Research Bulletin, 146, 171-184. https://doi.org/10.1016/j.brainresbull.2019.01.004 DOI: https://doi.org/10.1016/j.brainresbull.2019.01.004
Madhavi Sastry, G., Adzhigirey, M., Day, T., Annabhimoju, R., & Sherman, W. (2013). Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. Journal of Computer-Aided Molecular Design, 27(3), 221-234. https://doi.org/10.1007/s10822-013-9644-8 DOI: https://doi.org/10.1007/s10822-013-9644-8
Marucci, G., Buccioni, M., Ben, D. D., Lambertucci, C., Volpini, R., & Amenta, F. (2021). Efficacy of acetylcholinesterase inhibitors in Alzheimer’s disease. In Neuropharmacology, 190. https://doi.org/10.1016/j.neuropharm.2020.108352 DOI: https://doi.org/10.1016/j.neuropharm.2020.108352
Nemukhin, A. V, Grigorenko, B. L., Morozov, D. I., Kochetov, M. S., Lushchekina, S. V, & Varfolomeev, S. D. (2013). On quantum mechanical–molecular mechanical (QM/MM) approaches to model hydrolysis of acetylcholine by acetylcholinesterase. Chemico-Biological Interactions, 203(1), 51-56. DOI: https://doi.org/10.1016/j.cbi.2012.08.027
Nissink, J. W. M., Murray, C., Hartshorn, M., Verdonk, M. L., Cole, J. C., & Taylor, R. (2002). A new test set for validating predictions of protein–ligand interaction. Proteins: Structure, Function, and Bioinformatics, 49(4), 457-471. DOI: https://doi.org/10.1002/prot.10232
Oh, J. M., Kang, Y., Hwang, J. H., Park, J. H., Shin, W. H., Mun, S. K., Lee, J. U., Yee, S. T., & Kim, H. (2022). Synthesis of 4-substituted benzyl-2-triazole-linked-tryptamine-paeonol derivatives and evaluation of their selective inhibitions against butyrylcholinesterase and monoamine oxidase-B. International Journal of Biological Macromolecules, 217, 910-921. https://doi.org/10.1016/j.ijbiomac.2022.07.178 DOI: https://doi.org/10.1016/j.ijbiomac.2022.07.178
Oliyaei, N., Moosavi-Nasab, M., Tanideh, N., & Iraji, A. (2023). Multiple roles of fucoxanthin and astaxanthin against Alzheimer’s disease: Their pharmacological potential and therapeutic insights. In Brain Research Bulletin, 193, 11-21. https://doi.org/10.1016/j.brainresbull.2022.11.018 DOI: https://doi.org/10.1016/j.brainresbull.2022.11.018
Oyedele, A. Q. K., Adelusi, T. I., Ogunlana, A. T., Ayoola, M. A., Adeyemi, R. O., Babalola, M. O., Ayorinde, J. B., Isong, J. A., Ajasa, T. O., & Boyenle, I. D. (2023). Promising disruptors of p53-MDM2 dimerization from some medicinal plant phytochemicals: a molecular modeling study. Journal of Biomolecular Structure and Dynamics, 41(12), 5817-5826. https://doi.org/10.1080/07391102.2022.2097313 DOI: https://doi.org/10.1080/07391102.2022.2097313
Oyedele, A. Q. K., Ogunlana, A. T., Boyenle, I. D., Ibrahim, N. O., Gbadebo, I. O., Owolabi, N. A., Ayoola, A. M., Francis, A. C., Eyinade, O. H., & Adelusi, T. I. (2023). Pharmacophoric analogs of sotorasib-entrapped KRAS G12C in its inactive GDP-bound conformation: covalent docking and molecular dynamics investigations. Molecular Diversity, 27(4), 1795-1807. https://doi.org/10.1007/s11030-022-10534-1 DOI: https://doi.org/10.1007/s11030-022-10534-1
Pantsar, T., & Poso, A. (2018). Binding affinity via docking: fact and fiction. Molecules, 23(8), 1899. DOI: https://doi.org/10.3390/molecules23081899
Pasala, C., Katari, S. K., Nalamolu, R. M., Bitla, A. R., & Amineni, U. (2019). In silico probing exercises, bioactive-conformational and dynamic simulations strategies for designing and promoting selective therapeutics against Helicobacter pylori strains. Journal of Molecular Graphics and Modelling, 92, 167-179. https://doi.org/10.1016/j.jmgm.2019.07.015 DOI: https://doi.org/10.1016/j.jmgm.2019.07.015
Patridge, E., Gareiss, P., Kinch, M. S., & Hoyer, D. (2016). An analysis of FDA-approved drugs: natural products and their derivatives. Drug Discovery Today, 21(2), 204-207. DOI: https://doi.org/10.1016/j.drudis.2015.01.009
Pluta, R., Januszewski, S., & Czuczwar, S. J. (2021). Myricetin as a promising molecule for the treatment of post-ischemic brain neurodegeneration. In Nutrients, 13(2), 1-16). https://doi.org/10.3390/nu13020342 DOI: https://doi.org/10.3390/nu13020342
Podlewska, S., & Kafel, R. (2018). MetStabOn—online platform for metabolic stability predictions. International Journal of Molecular Sciences, 19(4), 1040. DOI: https://doi.org/10.3390/ijms19041040
Pritam, P., Deka, R., Bhardwaj, A., Srivastava, R., Kumar, D., Jha, A. K., Jha, N. K., Villa, C., & Jha, S. K. (2022). Antioxidants in Alzheimer’s Disease: Current Therapeutic Significance and Future Prospects. In Biology, 11(2). https://doi.org/10.3390/biology11020212 DOI: https://doi.org/10.3390/biology11020212
Ramezani, M., Darbandi, N., Khodagholi, F., & Hashemi, A. (2016). Myricetin protects hippocampal CA3 pyramidal neurons and improves learning and memory impairments in rats with Alzheimer’s disease. Neural Regeneration Research, 11(12), 1976. DOI: https://doi.org/10.4103/1673-5374.197141
Ramírez-Rendon, D., Passari, A. K., Ruiz-Villafán, B., Rodríguez-Sanoja, R., Sánchez, S., & Demain, A. L. (2022). Impact of novel microbial secondary metabolites on the pharma industry. In Applied Microbiology and Biotechnology, 106(5–6), 1855-1878. https://doi.org/10.1007/s00253-022-11821-5 DOI: https://doi.org/10.1007/s00253-022-11821-5
Rants, T. A., Westhuizen, C. J. Van Der, & Zyl, R. L. Van. (2022). Optimization of covalent docking for organophosphates interaction with Anopheles acetylcholinesterase. Journal of Molecular Graphics and Modelling, 110, 108054. https://doi.org/10.1016/j.jmgm.2021.108054 DOI: https://doi.org/10.1016/j.jmgm.2021.108054
Roleira, F. M. F., Tavares-Da-Silva, E. J., Varela, C. L., Costa, S. C., Silva, T., Garrido, J., & Borges, F. (2015). Plant derived and dietary phenolic antioxidants: Anticancer properties. In Food Chemistry, 183, 235-258. https://doi.org/10.1016/j.foodchem.2015.03.039 DOI: https://doi.org/10.1016/j.foodchem.2015.03.039
Rossi, A., Stagno, C., Piperno, A., Iraci, N., Panseri, S., Montesi, M., Feizi-Dehnayebi, M., Bassi, G., Di Pietro, M. L., & Micale, N. (2024). Anticancer activity and morphological analysis of Pt (II) complexes: Their DFT approach, docking simulation, and ADME-Tox profiling. Applied Organometallic Chemistry. https://doi.org/10.1002/aoc.7403 DOI: https://doi.org/10.1002/aoc.7403
Rostkowski, M., Olsson, M. H. M., Søndergaard, C. R., & Jensen, J. H. (2011). Graphical analysis of pH-dependent properties of proteins predicted using PROPKA. BMC Structural Biology, 11(1), 1-6. DOI: https://doi.org/10.1186/1472-6807-11-6
Schrödinger LigPrep, Schrödinger Release 2021-1, LLC, New York, NY (Schrödinger Release 2021-2). (2021). Schrödinger LLC.
Simunkova, M., Alwasel, S. H., Alhazza, I. M., Jomova, K., Kollar, V., Rusko, M., & Valko, M. (2019). Management of oxidative stress and other pathologies in Alzheimer’s disease. In Archives of Toxicology, 93(9), 2491-2513). https://doi.org/10.1007/s00204-019-02538-y DOI: https://doi.org/10.1007/s00204-019-02538-y
Song, X., Tan, L., Wang, M., Ren, C., Guo, C., Yang, B., Ren, Y., Cao, Z., Li, Y., & Pei, J. (2021a). Myricetin: A review of the most recent research. Biomedicine & Pharmacotherapy, 134, 111017.
Song, X., Tan, L., Wang, M., Ren, C., Guo, C., Yang, B., Ren, Y., Cao, Z., Li, Y., & Pei, J. (2021b). Myricetin: A review of the most recent research. In Biomedicine and Pharmacotherapy, 134. https://doi.org/10.1016/j.biopha.2020.111017 DOI: https://doi.org/10.1016/j.biopha.2020.111017
Srivastava, R. (2021). Theoretical Studies on the Molecular Properties, Toxicity, and Biological Efficacy of 21 New Chemical Entities. ACS Omega, 6(38), 24891-24901. https://doi.org/10.1021/acsomega.1c03736 DOI: https://doi.org/10.1021/acsomega.1c03736
Steinberg, G. M., Mednick, M. L., Maddox, J., Rice, R., & Cramer, J. (1975). Hydrophobic binding site in acetylcholinesterase. Journal of Medicinal Chemistry, 18(11), 1056-1061. DOI: https://doi.org/10.1021/jm00245a002
Sugimoto, H., Ogura, H., Arai, Y., Iimura, Y., & Yamanishi, Y. (2002). REVIEW-New drug and recent technique-research and development of donepezil hydrochloride, a new type of acetylcholinesterase inhibitor. In Japan Journal of Pharmacology, 89. DOI: https://doi.org/10.1254/jjp.89.7
Tamilselvan, M., Tamilanban, T., & Chitra, V. (2020). Unfolding remedial targets for Alzheimer’s disease. Research Journal of Pharmacy and Technology, 13(6), 3021-3027. DOI: https://doi.org/10.5958/0974-360X.2020.00534.X
Tavella, D., Ouellette, D. R., Garofalo, R., Zhu, K., Xu, J., Oloo, E. O., Negron, C., & Ihnat, P. M. (2022). A novel method for in silico assessment of Methionine oxidation risk in monoclonal antibodies: Improvement over the 2-shell model. PLoS ONE, 17. https://doi.org/10.1371/journal.pone.0279689 DOI: https://doi.org/10.1371/journal.pone.0279689
Tian, S., Jiang, L., Cui, X., Zhang, J., Guo, S., Li, M., Zhang, H., Ren, Y., Gong, G., Zong, M., Liu, F., Chen, Q., & Xu, Y. (2018). Engineering herbicide-resistant watermelon variety through CRISPR/Cas9-mediated base-editing. Plant Cell Reports, 37(9), 1353-1356. https://doi.org/10.1007/s00299-018-2299-0 DOI: https://doi.org/10.1007/s00299-018-2299-0
van Greunen, D. G., Cordier, W., Nell, M., van der Westhuyzen, C., Steenkamp, V., Panayides, J., & Riley, D. L. (2017). Targeting Alzheimer’s disease by investigating previously unexplored chemical space surrounding the cholinesterase inhibitor donepezil. European Journal of Medicinal Chemistry, 127, 671-690. https://doi.org/10.1016/j.ejmech.2016.10.036 DOI: https://doi.org/10.1016/j.ejmech.2016.10.036
van Greunen, D. G., Johan van der Westhuizen, C., Cordier, W., Nell, M., Stander, A., Steenkamp, V., Panayides, J.-L. L., Riley, D. L., van der Westhuizen, C. J., Cordier, W., Nell, M., Stander, A., Steenkamp, V., Panayides, J.-L. L., Riley, D. L., Johan van der Westhuizen, C., Cordier, W., Nell, M., Stander, A., … Riley, D. L. (2019). Novel N-benzylpiperidine carboxamide derivatives as potential cholinesterase inhibitors for the treatment of Alzheimer’s disease. European Journal of Medicinal Chemistry, 179, 680-693. https://doi.org/10.1016/j.ejmech.2019.06.088
van Greunen, D. G., van der Westhuizen, C. J., Cordier, W., Nell, M., Stander, A., Steenkamp, V., Panayides, J.-L. L., Riley, D. L., Johan van der Westhuizen, C., Cordier, W., Nell, M., Stander, A., Steenkamp, V., Panayides, J.-L. L., Riley, D. L., van der Westhuizen, C. J., Cordier, W., Nell, M., Stander, A., … Riley, D. L. (2019). Novel N-benzylpiperidine carboxamide derivatives as potential cholinesterase inhibitors for the treatment of Alzheimer’s disease. European Journal of Medicinal Chemistry, 179, 680-693. https://doi.org/10.1016/j.ejmech.2019.06.088 DOI: https://doi.org/10.1016/j.ejmech.2019.06.088
Wang, B., Zhong, Y., Gao, C., & Li, J. (2017a). Myricetin ameliorates scopolamine-induced memory impairment in mice via inhibiting acetylcholinesterase and down-regulating brain iron. Biochemical and Biophysical Research Communications, 490(2), 336-342.
Wang, B., Zhong, Y., Gao, C., & Li, J. (2017b). Myricetin ameliorates scopolamine-induced memory impairment in mice via inhibiting acetylcholinesterase and down-regulating brain iron. Biochemical and Biophysical Research Communications, 490(2), 336-342. https://doi.org/10.1016/j.bbrc.2017.06.045 DOI: https://doi.org/10.1016/j.bbrc.2017.06.045
Wessler, J. D., Grip, L. T., Mendell, J., & Giugliano, R. P. (2013). The P-glycoprotein transport system and cardiovascular drugs. Journal of the American College of Cardiology, 61(25), 2495-2502. DOI: https://doi.org/10.1016/j.jacc.2013.02.058
Xiong, G., Wu, Z., Yi, J., Fu, L., Yang, Z., Hsieh, C., Yin, M., Zeng, X., Wu, C., Lu, A., Chen, X., Hou, T., & Cao, D. (2021). ADMETlab 2.0: An integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Research, 49(W1), W5–W14. https://doi.org/10.1093/nar/gkab255 DOI: https://doi.org/10.1093/nar/gkab255
Yang, H., Lou, C., Sun, L., Li, J., Cai, Y., Wang, Z., Li, W., Liu, G., & Tang, Y. (2019). admetSAR 2.0: web-service for prediction and optimization of chemical ADMET properties. Bioinformatics, 35(6), 1067-1069. DOI: https://doi.org/10.1093/bioinformatics/bty707
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2025 Mojeed Ashiru, Rasheed Adewale Adigun, Musa Oladayo Babalola, Sherif Olabisi Ogunyemi, Idris Oladimeji Junaid, Maryam Titilayo Bello-Hassan, Mojisola Adebimpe Fategbe, Myah Grace Baker, Kazeem Adelani Alabi, Prince Ozioma Emmanuel, Mohammed O. Balogun

This work is licensed under a Creative Commons Attribution 4.0 International License.
Authors who publish with this journal agree to the following terms:
1) Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution License that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
2) Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the journal's published version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgement of its initial publication in this journal.
3) Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work.


