Purification and characterization of lipase from a new strain Staphylococcus argenteus MG2 (MTCC 12820)
DOI:
https://doi.org/10.14295/bjs.v3i5.547Keywords:
Staphylococcus argenteus MG2, lipase, purification, characterizationAbstract
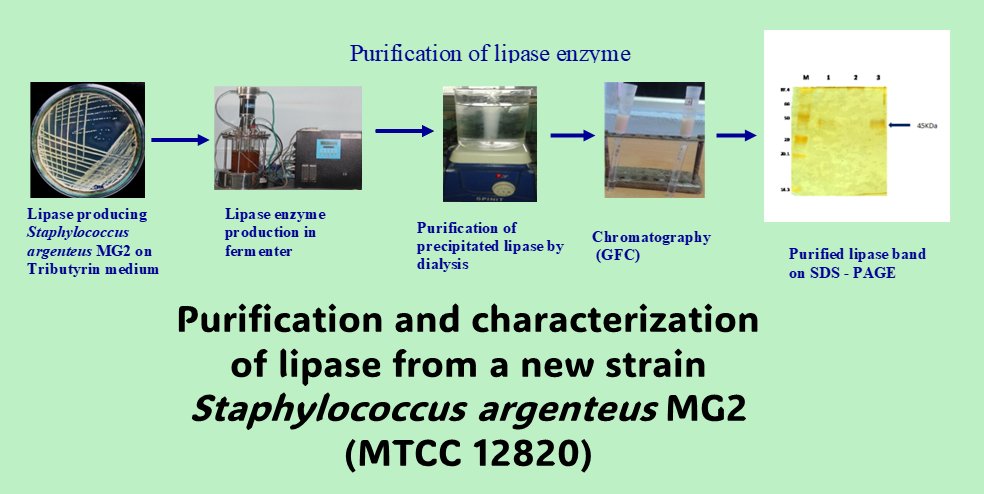
The lipase enzyme was isolated and purified from Staphylococcus argenteus MG2 (MTCC 12820) to homogeneity using ammonium sulphate precipitation followed by chromatographic techniques. This process resulted in a purification factor of 40.96-fold and a 26.25% recovery with a specific activity of 744.68 U mg-1. The molecular weight of the purified lipase was determined by SDS-PAGE to be 45 kDa. The Km and Vmax values of the purified lipase were calculated to be 4.95 mM and 79.36 µmol/min/mg-1, respectively. The maximum lipase activity was observed at pH 7.0 and 30 ºC with 100% stability, and it was also found to be stable in a broad range of pH (5-12) and temperature (30-90 ºC). The enzymatic activity of this Staphylococcal lipase was increased by Ca2+ to 105.71% at a concentration of 1 mM CaCl2. Additionally, it exhibited marked stability and activity in organic solvents. In the presence of 1% SDS surfactant, it retained 85.16% residual activity, while the metal chelator EDTA (inhibitor) reduced the lipase activity to 83.87% residual activity at a concentration of 1% w/v. This alkali-stable and thermo-stable lipase can be exploited by extending its use in the preparation of detergents and in various industrial and biotechnological applications.
References
Aires-Barros, M. R., Taipa, M. A., & Cabral, J. M. S. (1994). Isolation and purification of lipases. In: Lipases: their structure, biochemistry and application. Great Britain: Cambridge University Press, Eds. Wooley, P., & Petersen, S. B., 234-270 p.
Bacha, A. B., Al-Assaf, A., Moubayed, N. M., & Abid, I. (2018). Evaluation of a novel thermo-alkaline Staphylococcus aureus lipase for application in detergent formulations. Saudi Journal of Biological Sciences, 25(3), 409-417. https://doi.org/10.1016%2Fj.sjbs.2016.10.006 DOI: https://doi.org/10.1016/j.sjbs.2016.10.006
Beisson, F., Tiss, A., Riviere, C., & Verger, R. (2000). Methods for lipase detection and assay: a critical review. European Journal of Lipid Science and Technology, 102, 133-153. https://doi.org/10.1002/(SICI)1438-9312(200002)102:2%3C133::AID-EJLT133%3E3.0.CO;2-X DOI: https://doi.org/10.1002/(SICI)1438-9312(200002)102:2<133::AID-EJLT133>3.3.CO;2-O
Brune, A. K., & Gotz, F. (1992). In: Microbial degradation of natural products. (ed. Winkelmann, G). VCH, Weinheim, 243-263 p.
Castro-Ochoa, L. D., Rodriguez-Gomez, C., Valerio-Alfaro, G., & Ros, R. O. (2005). Screening, purification and characterization of the thermo alkalophilic lipase produced by Bacillus thermoleovorans CCR11. Enzyme and Microbial Technology, 37(6), 648-654. https://doi.org/10.1016/j.enzmictec.2005.06.003 DOI: https://doi.org/10.1016/j.enzmictec.2005.06.003
Chartrain, M., Katz, L., Marcin, C., Thien, M., Smith, S., Fisher, F., Goklen, K., Salmon, P., Brix, T., Price, K., & Greasham, R. (1993). Purification and characterization of a novel bioconverting lipase from Pseudomonas aeruginosa MB 5001. Enzyme and Microbial Technology, 15(7), 575-580. https://doi.org/10.1016/0141-0229(93)90019-X DOI: https://doi.org/10.1016/0141-0229(93)90019-X
Chen, S. J., Cheng, C. Y., & Chen, T. L. (1998). Production of an alkaline lipase by Acinetobacter radioresistens. Journal of Fermentation and Bioengineering, 86(3), 308-312. https://doi.org/10.1016/S0922-338X(98)80135-9 DOI: https://doi.org/10.1016/S0922-338X(98)80135-9
Cherif, S., Mnif, S., Hadrich, F., Abdelkafi, S., & Sayadi, S. (2011). A newly high alkaline lipase: an ideal choice for application in detergent formulations. Lipids in Health and Disease, 10, 1-8. https://doi.org/10.1186/1476-511X-10-221 DOI: https://doi.org/10.1186/1476-511X-10-221
Dharmsthiti, S., Pratuangdejkul, J., Theeragool, G., & Luchai, S. (1998). Lipase activity and gene cloning of Acinetobacter calcoaceticus LP009. The Journal of General and Applied Microbiology, 44(2), 139-145. https://doi.org/10.2323/jgam.44.139 DOI: https://doi.org/10.2323/jgam.44.139
Farrell, A. M., Foster, T. J., & Holland, K. T. (1993). Molecular analysis and expression of the lipase of Staphylococcus epidermidis. Microbiology, 139(2), 267-277. https://doi.org/10.1099/00221287-139-2-267 DOI: https://doi.org/10.1099/00221287-139-2-267
Ghori, M. I., Iqbal, M. J., & Hameed, A. (2011). Characterization of a novel lipase from Bacillus sp. isolated from tannery wastes. Brazilian Journal of Microbiology, 42(1), 1-11. https://doi.org/10.1590%2FS1517-83822011000100003 DOI: https://doi.org/10.1590/S1517-83822011000100003
Godfredson, S. E. (1990). In: Microbial enzymes and biotechnology. (eds Fogarty WM and Kelly ET). Elsevier. Applied Sciences, The Netherlands, 255-273 p.
Golani, M., Hajela, K., & Pandey, G. P. (2019). Isolation and identification of a novel lipase producing Staphylococcus argenteus MG2 bacterium from oil spilled soil. International Journal of Advance and Innovative Research, 6(1), 20-30. https://iaraedu.com/about-journal/ijair-volume-6-issue-1-xiv-january-march-2019.php
Gotor, V. (2002). Lipases and (R)-oxynitrilases: useful tools in organic synthesis. Journal of Biotechnology, 96(1), 35-42. https://doi.org/10.1016/s0168-1656(02)00035-4 DOI: https://doi.org/10.1016/S0168-1656(02)00035-4
Gotz, F., Popp, F., Korn, E., & Schleifer, K. H. (1985). Complete nucleotide sequence of the lipase gene from Staphylococcus hyicus cloned in Staphylococcus carnosus. Nucleic Acids Research, 13(16), 5895-5906. https://doi.org/10.1093/nar/13.16.5895 DOI: https://doi.org/10.1093/nar/13.16.5895
Haki, G. D., & Rakshit, S. K. (2003). Developments in industrially important thermostable enzymes: a review. Bioresource Technology, 89(1), 17-34. https://doi.org/10.1016/S0960-8524(03)00033-6 DOI: https://doi.org/10.1016/S0960-8524(03)00033-6
Handelsman, T., & Shoham, Y. (1994). Production and characterization of an extracellular thermostable lipase from a thermophilic Bacillus sp. The Journal of General and Applied Microbiology, 40(5), 435-443. https://doi.org/10.2323/jgam.40.435 DOI: https://doi.org/10.2323/jgam.40.435
Hiol, A., Jonzo, M. D., Rugani, N., Druet, D., Sarda, L., & Comeau, L. C. (2000). Purification and characterization of an extracellular lipase from a thermophilic Rhizopus oryzae strain isolated from palm fruit. Enzyme Microbial Technology, 26(5-6), 421-430. https://doi.org/10.1016/s0141-0229(99)00173-8 DOI: https://doi.org/10.1016/S0141-0229(99)00173-8
Horchani, H., Mosbah, H., Salem, N.B., Gargouri, Y., & Sayari, A. (2009). Biochemical and molecular characterization of a thermoactive, alkaline and detergent-stable lipase from a newly isolated Staphylococcus aureus strain. Journal of Molecular Catalysis B: Enzymatic, 56(4), 237-245. http://dx.doi.org/10.1016/j.molcatb.2008.05.011 DOI: https://doi.org/10.1016/j.molcatb.2008.05.011
Illanes, A. (1999). Stability of biocatalysts. Electronic Journal of Biotechnology, 2(1), 7-15. http://dx.doi.org/10.4067/S0717-34581999000100001 DOI: https://doi.org/10.2225/vol2-issue1-fulltext-2
Jaeger, K E., Dijkstra, B. W., & Reetz, M. T. (1999). Bacterial biocatalysts: molecular biology, three-dimensional structures, and biotechnological applications of lipases. Annual Reviews in Microbiology, 53(1), 315-351. https://doi.org/10.1146/annurev.micro.53.1.315 DOI: https://doi.org/10.1146/annurev.micro.53.1.315
Jaeger, K-E., Ransac, S., Dijkstra, B.W., Colson, C., Heuvel, M., & Van, M. O. (1994). Bacterial lipases. FEMS Microbiol Reviews, 15, 29-63. https://doi.org/10.1111/j.1574-6976.1994.tb00121.x DOI: https://doi.org/10.1111/j.1574-6976.1994.tb00121.x
Junxin, Z., Maomao, M., Zheling, Z., Ping, Y., Deming, G. & Shuguang, D. (2021). Production, purification and biochemical characterization of a novel lipase from a newly identified lipolytic bacterium Staphylococcus caprae NCU S6. Journal of Enzyme Inhibition and Medicinal Chemistry, 36(1), 249-257. https://doi.org/10.1080/14756366.2020.1861607 DOI: https://doi.org/10.1080/14756366.2020.1861607
Kanwar, L., Gogoi, B. K., & Goswami, P. (2002). Production of a Pseudomonas lipase in n-alkane substrate and its isolation using an improved ammonium sulfate precipitation technique. Bioresource Technology, 84(3), 207-211. https://doi.org/10.1016/s0960-8524(02)00061-5 DOI: https://doi.org/10.1016/S0960-8524(02)00061-5
Kim, S. S., E. K., & Rhee, J. S. (1996). Effects of growth rate on the production of Pseudomonas fluorescens lipase during the fed-batch cultivation of Escherichia coli. Biotechnology Progress, 12(5), 718-722. https://doi.org/10.1021/bp960047h DOI: https://doi.org/10.1021/bp960047h
Kulkarni, N., & Gadre, R.V. (2002). Production and properties of an alkaline, thermophilic lipase from Pseudomonas fluorescens NS2W. Journal of Industrial Microbiology and Biotechnology, 28, 344-348. https://doi.org/10.1038/sj/jim/7000254 DOI: https://doi.org/10.1038/sj.jim.7000254
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227(5259), 680-685. https://doi.org/10.1038/227680a0 DOI: https://doi.org/10.1038/227680a0
Lee, C. Y., & Iandolo, J. J. (1986). Lysogenic conversion of Staphylococcal lipase is caused by insertion of the bacteriophage L54a genome into the lipase structural gene. Journal of Bacteriology, 166(2), 385-391. https://doi.org/10.1128/jb.166.2.385-391.1986 DOI: https://doi.org/10.1128/jb.166.2.385-391.1986
Linefield, W. M., Barauskas, R. A., Siviri, L., Serota, S., & Steveson, S. W. (1990). Enzymatic fat hydrolysis and synthesis. Journal of the Amirican Oil Chemists’ Society, 61, 191-195. https://doi.org/10.1007/BF02678767 DOI: https://doi.org/10.1007/BF02678767
Longshaw, C. M., Farrell, A. M., Wright, J. D., & Holland, K. T. (2000). Identification of a second lipase gene, gehD, in Staphylococcus epidermidis: comparison of sequence with those of other staphylococcal lipases. Microbiology, 146(6), 1419-1427. https://doi.org/10.1099/00221287-146-6-1419 DOI: https://doi.org/10.1099/00221287-146-6-1419
Macedo, G. A., Park, Y. K., & Pastore, G. M. (1997). Partial purification and characterization of an extracellular lipase from a newly isolated strain of Geotrichum sp. Brazilian Journal of Microbiology, 39, 687-692.
Mosbah, H., Sayari, A., Mejdoub, H., Dhouib, H., & Gargouri, Y. T. (2005). Biochemical and molecular characterization of Staphylococcus xylosus lipase. Biochimica et Biophysica Acta, 1723, 282-291. https://doi.org/10.1016/j.bbagen.2005.03.006 DOI: https://doi.org/10.1016/j.bbagen.2005.03.006
Nawani, N., & Kaur, J. (2000). Purification, characterization and thermostability of lipase from a thermophilic Bacillus sp.J33. Molecular and Cellular Biochemistry, 206, 91-96. https://doi.org/10.1023/a:1007047328301 DOI: https://doi.org/10.1023/A:1007047328301
Nishio, T., Chikano, T., & Kamimura, M. (1987). Purification and some properties of lipase produced by Pseudomonas fragi22.39 B. Agricultural and Biological Chemistry, 51(1), 181-187. https://doi.org/10.1080/00021369.1987.10867979 DOI: https://doi.org/10.1080/00021369.1987.10867979
Oh, B. C., Kim, H. K., Lee, J. K., Kang, S. C., & Oh, T. K. (1999). Staphylococcus haemolyticus lipase: biochemical properties, substrate specificity and gene cloning. FEMS Microbiology Letters, 179(2), 385-392. https://doi.org/10.1111/j.1574-6968.1999.tb08753.x DOI: https://doi.org/10.1111/j.1574-6968.1999.tb08753.x
Pandey, A., Benjamin, S., Soccol, C.R., Nigam, P., Krieger, N., & Soccol, V. T. (1999). The realm of microbial lipases in biotechnology. Biotechnology and Applied Biochemistry, 29(2), 119-131. PMID: 10075908 . DOI: https://doi.org/10.1111/j.1470-8744.1999.tb00541.x
Rathi, P., Saxena, R. K., & Gupta, R. (2001). A novel alkaline lipase from Burkholderia cepacia for detergent formulation. Process Biochemistry, 37(2), 187-192. http://dx.doi.org/10.1016/S0032-9592(01)00200-X DOI: https://doi.org/10.1016/S0032-9592(01)00200-X
Saxena, R. K., Ghosh, P. K., Gupta, R., Davidson, W. S., Bradoo, S., & Gulati, R. (1999). Microbial lipases: Potential biocatalysts for the future industry. Current Science, 77(1), 101-115.
Sayari, A., Agrebi, N., Jaoua, S., & Gargouri, Y. (2001). Biochemical and molecular characterization of Staphylococcus simulans lipase. Biochimie, 83(9), 863-871. https://doi.org/10.1016/s0300-9084(01)01327-x DOI: https://doi.org/10.1016/S0300-9084(01)01327-X
Sigman, D. S., & Mooser, G. (1975). Chemical studies of enzyme active sites. Annual Review of Biochemistry, 44, 889-931. https://doi.org/10.1146/annurev.bi.44.070175.004325 . DOI: https://doi.org/10.1146/annurev.bi.44.070175.004325
Simons, J. W. F., van Kampen, M. D., Riel, S., Gotz, F., Egmond, M. R., & Verheij, H. M. (1998). Cloning, purification and characterization of the lipase from Staphylococcus epidermidis: Comparison of the substrate selectivity with those of other microbial lipases. European Journal of Biochemistry, 253(3), 675-683. https://doi.org/10.1046/j.1432-1327.1998.2530675.x DOI: https://doi.org/10.1046/j.1432-1327.1998.2530675.x
Sirisha, E., Lakshmi, P., & Lakshmi, N. M. (2017). Purification and characterization of extracellular lipase from Staphylococcus epidemidis (MTCC10656). Journal of Applied Pharmaceutical Science, 7(01), 57-63. http://dx.doi.org/10.7324/JAPS.2017.70108 DOI: https://doi.org/10.7324/JAPS.2017.70108
Talon, R., Dublet, N., Montel, M. C., & Cantonnet, M. (1995). Purification and characterization of extracellular Staphylococcus warneri lipase. Current Microbiology, 30, 11-16. https://doi.org/10.1007/BF00294517 DOI: https://doi.org/10.1007/BF00294517
Talon, R., Marie-Christine, M., & Jean- Louis, B. (1996). Production of flavor esters by lipases of Staphylococcus warneri and Staphylococcus xylosus. Enzyme and Microbial Technology, 19(8), 620-622. https://doi.org/10.1016/S0141-0229(96)00075-0 DOI: https://doi.org/10.1016/S0141-0229(96)00075-0
Tiesinga, J. J. W., Pouderoyen, V. G., Nardini, M., Ransac, S., & Dijkstra, B. W. (2007). Structural basis of phospholipase activity of Staphylococcus hyicus lipase. Journal of Molecular Biology, 371(2), 447-456. https://doi.org/10.1016/j.jmb.2007.05.041 DOI: https://doi.org/10.1016/j.jmb.2007.05.041
Van Oort, M. G., Deveer, A. M., Dijkman, R., Tjeenk, M. L., Verheij, H. M., De Haas, G. H., Wenzig, E., & Goetz, F. (1989). Purification and substrate specificity of Staphylococcus hyicus lipase. Biochemistry, 28(24), 9278-9285. https://doi.org/10.1021/bi00450a007 DOI: https://doi.org/10.1021/bi00450a007
VanKampen, M. D., Rosenstein, R., Gotz, F., & Egmond, M. R. (2001). Cloning, purification and characterization of Staphylococcus warneri lipase 2. Biochemica et Biophysica Acta (BBA) – Protein Structure and Molecular Enzymology, 1544(1-2), 229-241. https://doi.org/10.1016/s0167-4838(00)00224-7 DOI: https://doi.org/10.1016/S0167-4838(00)00224-7
Verma, M., Azmi, W., & Kanwar, S. (2008). Microbial lipases: at the interface of aqueous and non-aqueous media: a review. Acta Microbiologica et Immunologica Hungarica, 55(3), 265-294. https://doi.org/10.1556/amicr.55.2008.3.1 DOI: https://doi.org/10.1556/AMicr.55.2008.3.1
Woolley, P. (1994). Lipases; their structure, biochemistry and application. In: Sequence analysis of lipases, esterases and related proteins, Cambridge University Press, 27-28 p. https://cir.nii.ac.jp/crid/1570291224803998080
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2024 Mahima Golani, Krishnan Hajela

This work is licensed under a Creative Commons Attribution 4.0 International License.
Authors who publish with this journal agree to the following terms:
1) Authors retain copyright and grant the journal right of first publication with the work simultaneously licensed under a Creative Commons Attribution License that allows others to share the work with an acknowledgement of the work's authorship and initial publication in this journal.
2) Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the journal's published version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgement of its initial publication in this journal.
3) Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work.


